Phytochemistry & Biodiscovery
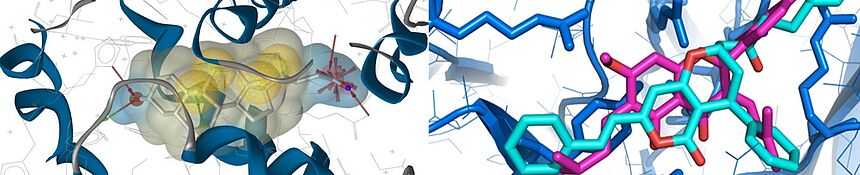
To increase the success rate in finding bioactive natural material, we implement computational and biochemometric techniques in pharmacognostic workflows. For instance, we apply pharmacophore modelling and molecular docking for virtual screening of natural product molecular databases, as well as chemometric approaches, e.g. using heterocovariance analyses.
Predicted ligand target interactions are used to provide insights into the molecular mechanism of identified bioactive compounds and to unravel the polypharmacological complexity of botanicals.
Selected Papers:
Wasilewicz A, Zwirchmayr J, Kirchweger B, Bojkova D, Cinatl J Jr, Rabenau HF, Rollinger JM, Beniddir MA, Grienke U. Discovery of anti-SARS-CoV-2 secondary metabolites from the heartwood of Pterocarpus santalinus using multi-informative molecular networking. Front. Mol Biosci. 2023 Jun 6;10:1202394. doi:10.3389/fmolb.2023.1202394.
Zwirchmayr J, Schachner D, Grienke U, Rudžionytė I, de Martin R, Dirsch VM, Rollinger JM. Biochemometry identifies suppressors of pro-inflammatory gene expression in Pterocarpus santalinus heartwood. Phytochemistry. 2023 Aug; 212:113709. doi: 10.1016/j.phytochem.2023.113709.
Wasilewicz A, Kirchweger B, Bojkova D, Abi Saad M, Langeder J, Bütikofer M, Adelsberger S, Grienke U, Cinatl J, Petermann O, Scapozza L, Orts J, Kirchmair J, Rabenau, H, Rollinger JM. Identification of natural products inhibiting SARS-CoV-2 by targeting the viral proteases: a combined in silico and in vitro approach. J. Nat. Prod. 2023, 86 (2), 264-275, 10.1021/acs.jnatprod.2c00843 ACS Editors’ Choice
Langeder J, Döring K, Schmietendorf H, Grienke U, Schmidtke M, Rollinger JM. 1H NMR based biochemometric analysis of Morus alba extracts towards a multipotent herbal anti-infective. J. Nat. Prod. 2023, 86 (1), 8-17. 10.1021/acs.jnatprod.2c00481 ACS Author Choice
Kirchweger B, Wasilewicz A, Fischhuber K, Tahir A, Chen Y, Heiss EH, Langer T, Kirchmair J, Rollinger JM. In silico and in vitro approach to assess direct allosteric AMPK activators from nature. Planta Med. 2022, 88(9-10):794-804. 10.1055/a-1797-3030
Zwirchmayr J, Grienke U, Hummelbrunner S, Seigner J, de Martin R, Dirsch VM, Rollinger JM. A biochemometric approach for the identification of in vitro anti-inflammatory constituents in masterwort. Biomolecules 2022; 10:679. 10.3390/biom10050679
Kirchweger B, Rollinger JM. A strength-weaknesses-opportunities-threats (SWOT) analysis of cheminformatics in natural product research. Progress in the Chemistry of Organic Natural Products 2019; 110: 239-271. 10.1007/978-3-030-14632-0_7
Grienke U, Foster PA, Zwirchmayr J, Tahir A, Rollinger JM, Mikros E. 1H NMR-MS-based heterocovariance as a drug discovery tool for fishing bioactive compounds out of a complex mixture of structural analogues. Scientific Reports 2019; 9: 1-10. 10.1038/s41598-019-47434-8